Written by SynBio CDT students Claire Noble and Harry Thompson.
Do we have any chance of designing new ribosomes from scratch? Maybe not just yet, but that doesn’t mean Jon Bath, from the University of Oxford, isn’t getting started. While DNA origami hasn’t always been as glamorous as the world of protein design, that doesn’t mean there isn’t lots of exciting potential for new, DNA-based biomachinery.
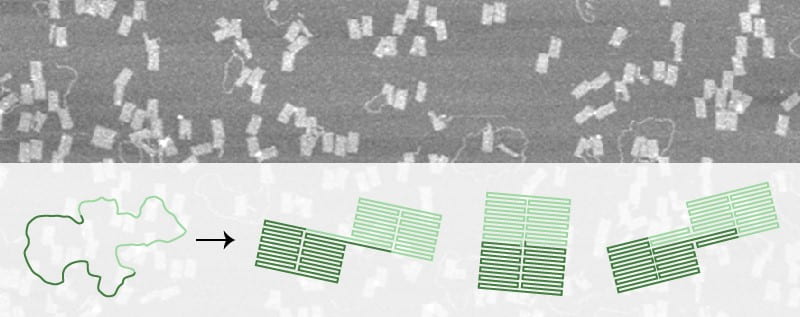
The relatively simple nature of DNA folding based on base pairing has allowed for the construction of intricate and beautiful DNA structures. However, the field of designing DNA structure towards novel functionality is still being explored. In the past, DNA has been shown to be capable of moving along short tracks and assembling simple polymers in a directed way. Jon Bath is seeking to gain a deeper fundamental understanding of what dictates higher level folding in DNA origami, so that more complex designs can be attempted. He is making use of comparatively ‘simple’ DNA structures, with uncommon motif’s such as T-junctions, to try and elucidate the mechanisms behind self-assembly of complex origami.
By increasing our understanding of how DNA folds, design principles can then be applied towards constructions of functional origamis, of which there have been relatively few examples so far. A brave new world of DNA templated chemistry and molecular motors awaits!